DrugSig: drug induced gene signature for drug repositioning
User's Guide
Browse Search Tools Disclaimer
Over the past decades, to bring a new drug to the market often takes billions of investment dollars and an average about 9-12 years. De novo drug discovery has grown to be time-consuming and costly. In light of these challenges, drug repositioning, which concerns the detection and development of new clinical indications for those existing drugs has emerged as an increasingly important strategy for the new drug discovery.
Historically, the discovery of new uses of old drugs is mostly through serendipity or resulted from a better understanding of the drugs’ mechanism of action. The efficacy of these methods is very low. With drug-related data growth and open data initiatives, a set of new repositioning strategies and techniques has emerged with integrating data from various sources, like pharmacological, genetic, chemical or clinical data. These methods can accumulate evidence supporting discovery of new uses or indications of existing drugs. However, currently existing strategies strongly rely on separated or individual experimental data, and resulted in inefficient outputs. The construction of an integrating drug-related database is a must.
We collected the gene expression data scattered in the separated databases to develop the drug-related database.
Workflow
We searched the scientific literature by keywords “human cell AND treatment AND ("gene signature" OR "expression profile") AND (genechip OR microarray OR "gene expression") AND English [la]” from PubMed. Finally, we constructed the drug signatures from papers or related records in GEO database.
After we got the raw data, we first read the data via rma method of affy package in BioConductor. We constructed the drug induced signatures (top 500 up-regulated and down-regulated probes) using two kinds of approaches depending on the type of raw data: we first read the data via rma method of affy package in BioConductor, then a) if the replicates < 3 in microarray experiments, we compute the signatures by simple fold changes (FC > 2.0 or FC < 0.5 ); b) if the replicates >=3, we compute the signatures by linear models implemented in Limma package of BioConductor program (FC > 2.0 or FC < 0.5 and P value < 0.01). In addition, if the number of calculated differential expressed probes < 500, we selected all differential expressed probes as signatures.
The constructed database can server as a tool to quicken drug repositioning.
Browse
The database browse interface provides the users with a function of navigating the entire database.
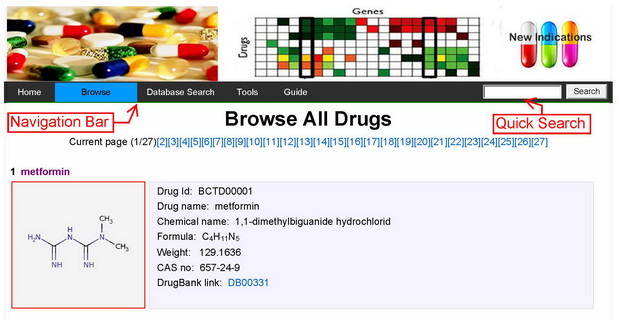 |
Figure 1The database browse interface provides the users with a function of navigating the entire database. |
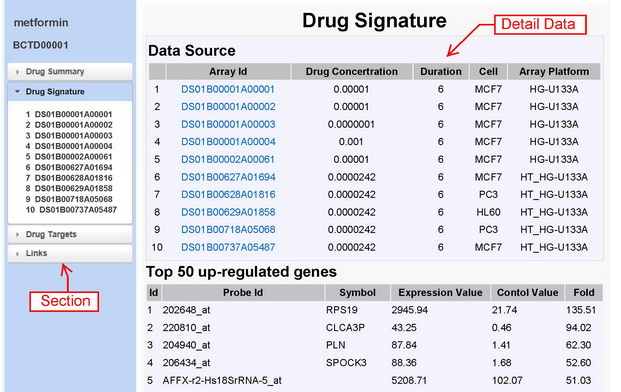 |
Figure 2A detail page with four sections: drug summary, drug signature, drug targets and links. Drug summary section consists of drug name, chemical name, formula, CAS no, description and drug indications. In addition, it also provides a link to DrugBank. Drug signature section demonstrates its common signatures which are comprised of top 50 up-regulated and down-regulated genes and its data source. For each microarray, there is a page to display its signatures. The drug targets section consists of the drug targets and their expression value in cells treated by the drug and other drugs. |
Search
We classified search into simple search and complex search.
 |
Figure 3The Search interface can be used to retrieve specific information using either a quick or advanced option.Simple search allows users to search database based on keywords like "drug name". Complex search allows users restrict the search to a particular field descriptor or a combination of varied field description. |
All searches are case insensitive. A complete list of the field descriptors and their description is given below:
DESCRIPTORS |
DESCRIPTION |
| Drug Id | Drug Id, such as BCTD00001
|
| Drug Name | Drug name, such as metformin
|
| DrugBank Id | DrugBank Id, such as DB00331
|
| Disease | Disease abbreviation, such as NIDDM |
| Target | Drug target symbol, for example:PRKAB1 |
| Signature Symbol | Gene symbol of drug signature, for example:CXCL11 |
Tools
Tools of drug repurposing implemented in DrugSig consist of signature based and target based drug repositioning functions.
The signature based drug repositioning function provides an interface to input user’s gene list to compute against DrugSig.
 |
Figure 4An interface for inputting user's gene list and searching task history by user's email. |
 |
Figure 5An interface for signature based computing. |
After submitting the computing request, a function will calculate the scores compute the scores. Once the computing finished, DrugSig will sent a notice email to user.
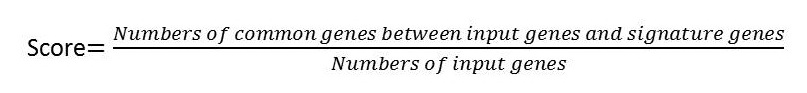 |
The results can be accessed later from the email or by searching the task history with user’s email address (Figure 4). The computing result contains queried gene list, top 50 score drugs produced reverse gene list and top 50 score drugs produced similar gene list.
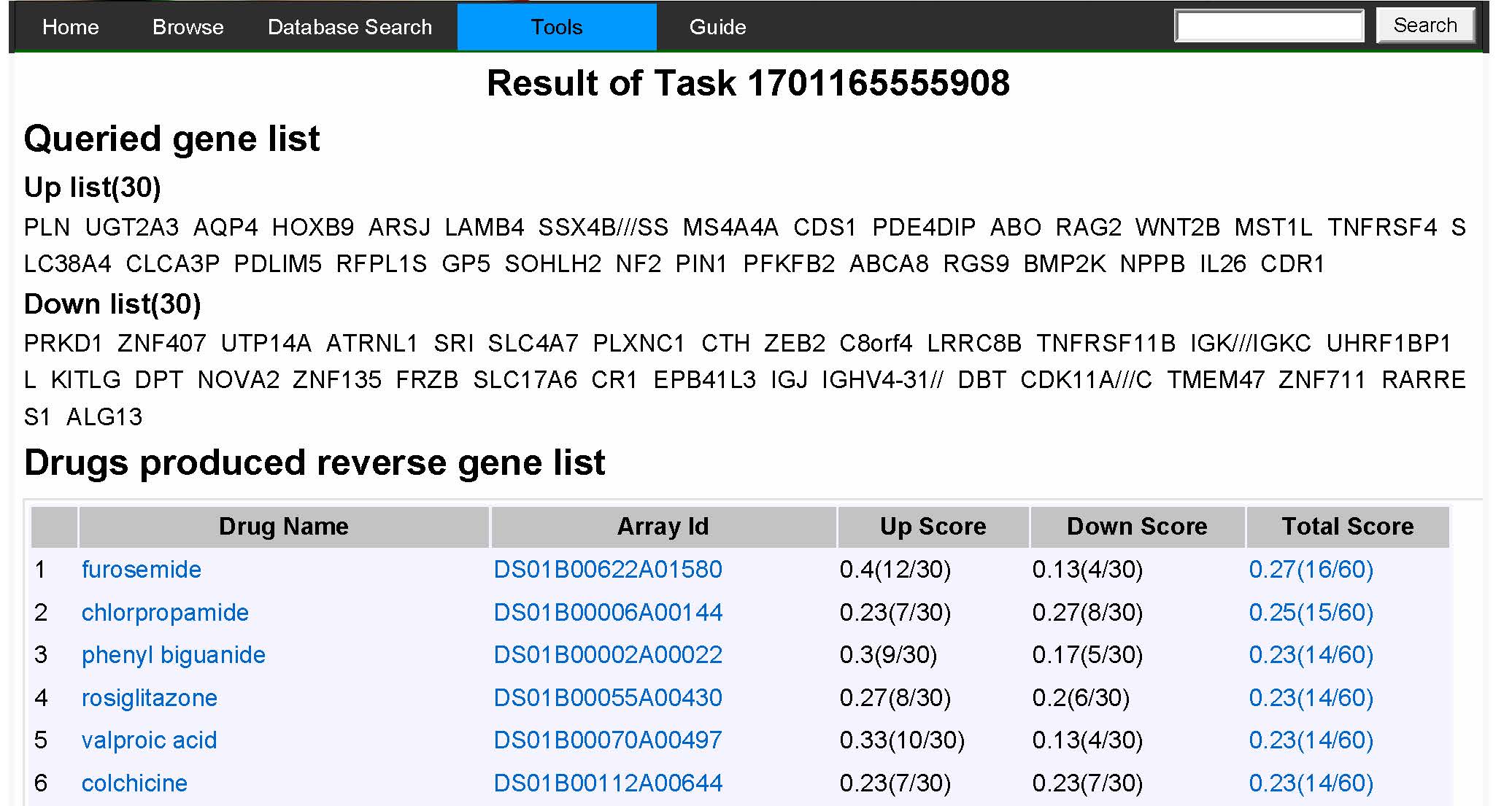 |
Figure 6A result for signature based computing showes top scores drugs produced reverse gene list. |
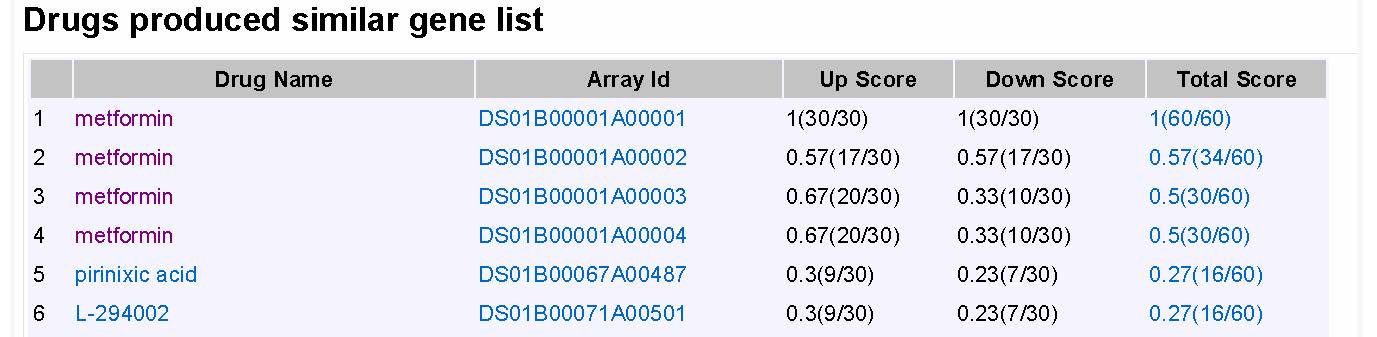 |
Figure 7A result for signature based computing showes top scores drugs produced similar gene list. |
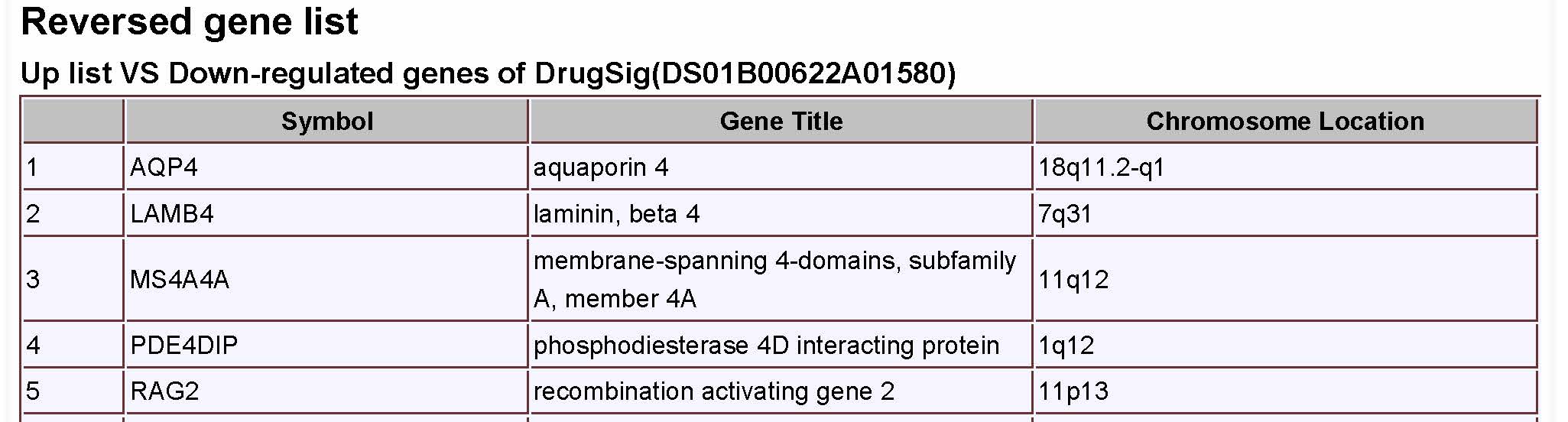 |
Figure 8A page showes a reverse gene list for the score in the result page. |
 |
Figure 9A page showes a similar gene list for the score in the result page. |
The target based drug repositioning function provides an interface to explore the specified target and its targeting drugs, as well as expression of target gene in cells after treated by drugs. The targeting drugs may have similar indications for drug repurposing. The expression level of target partly reflects the potential of the drug which inhibits or enhances the target.
 |
Figure 10An inferface for target based drug repositioning function. |
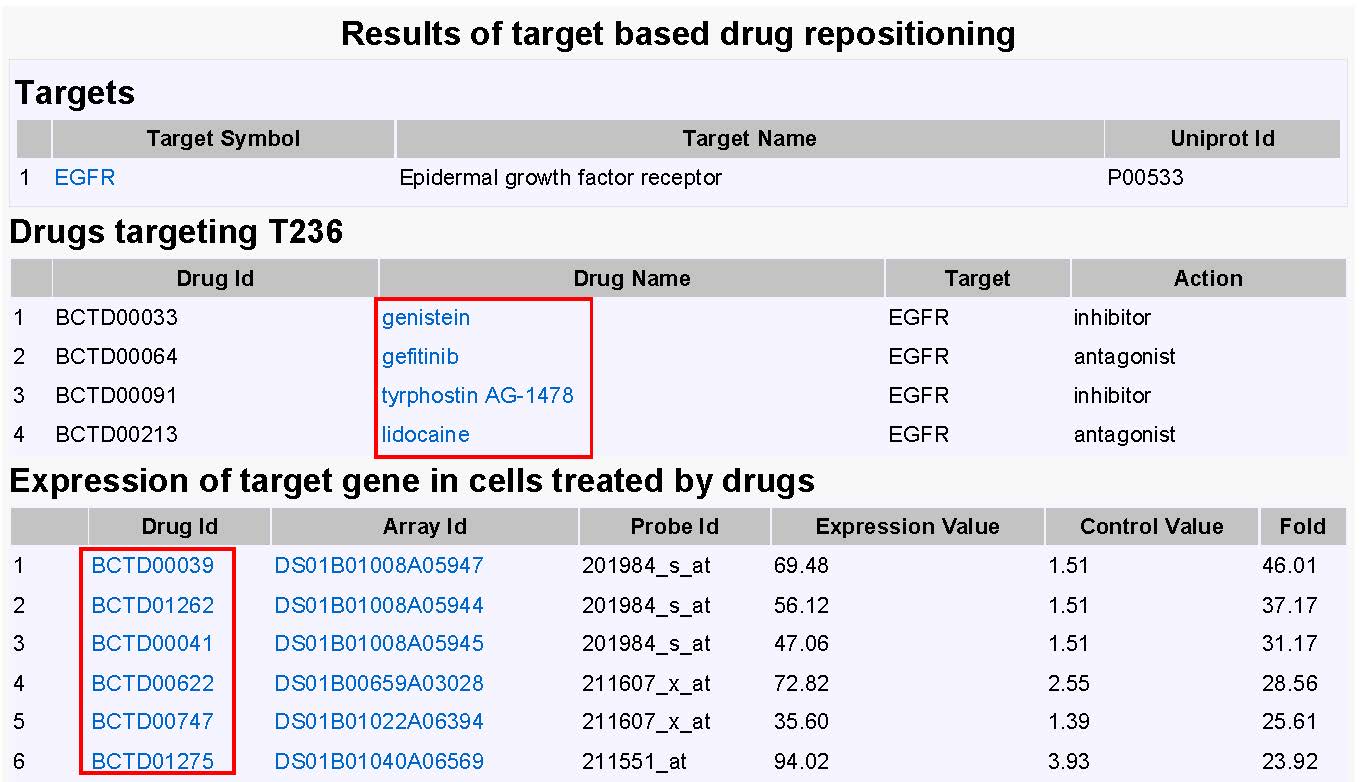 |
Figure 11The result of target based drug repositioning. |
Disclaimer
The authors do not assume any responsibility for losses of any kind incurred by use of this database.
