EnzyBase: A database for enzybiotic studies
Enzybiotics in EnzyBase refer to bacterial cell wall-degrading enzymes including lysins, bacteriocins, autolysins, and lysozymes. The most important characteristics of enzybiotics are a novel mode of antibacterial action, different from those typical of antibiotics, and the capacity to kill antibiotic-resistant bacteria. Another significant feature of some enzybiotics is the low probability of developing bacterial resistance.
EnzyBase (Collection of enzybiotics) has been created with an objective to provide a useful resource for study of enzybiotics. This manually curated database currently holds 1144 validated enzybiotics sequences from 216 nature sources. Information related to protein name, protein full name, producer organism, simple function annotation and protein sequence, domains, 3D-structure and regarding references, target organisms with MIC values, and links to external databases like UniProt, PDB and InterPro. PubMed links are included here.
A complete documentation of the database features and tools can be accessed from the Guide. The database will be updated monthly with additional sequences. This work has been partly funded by the Major scientific and technological specialized project of China for ‘significant new formulation of new drugs’ (grant 2008ZX09101-032) and ‘Yangtze river delta’ joint scientific and technological project of China (grant 10495810600).
People involved:
Dr. Qingshan Huang |
|
Mr. Hongyu Wu |
|
Mr. Jinjiang Huang |
|
Dr. Hairong Lu |
|
Mr. Guodong Li |
|
Publication:
1. Wu H, Lu H, Huang J, Li G, Huang Q (2012) EnzyBase: a novel database for enzybiotic studies. BMC Microbiol 12: 54. |
| Monthly Enzybiotic |
Feb. 2012
|
|
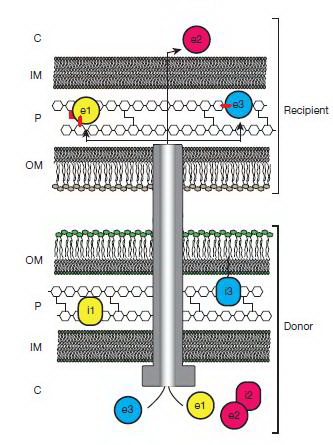
A model forT6S-catalysed translocation of effectors to the periplasm of recipient bacteria
Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD: Type VI secretion delivers bacteriolytic effectors to target cells. Nature 2011, 475:343-347. |
|